In his new highly technical paper, Mik Andersen presents his findings on the DNA crystals observed in the Pfizer vaccine: their comparison with the scientific literature; their implications in the development of circuits, transistors, electronic devices; their implications in the intracorporeal network of nano-communications and nano-devices; the development of computer systems, etc. In addition, it explains, first, how graphene and hydrogel are related to synthetic DNA structures and, second, the processes of graphene fusion with DNA. Finally, he presents an analysis and update of his research – taking into account all these recent advances. In conclusion, Mik Andersen considers this probably one of the most fundamental articles on his Corona2Inspect blog.
Microscopic images taken by Dr. (Campra, P. 2021) continue to provide valuable information for the elucidation of the contents and materials of vaccine vials. On this occasion, a key piece of evidence was discovered that would confirm that the Pfizer Comirnaty™ vaccine has DNA crystal self-assembly technology, consistent with the previous article on DNA-Origami self-assembly evidence. This discovery was made possible by comparing the images of Dr. Campra and Ricardo Delgado (Delgado, R. 2022) with the scientific literature, particularly the work by (Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018) entitled “Modulation of DNA crystal self-assembly with rationally designed agents”/“Modulation of DNA crystal self-assembly with rationally designed agents”. This discovery facilitates the understanding of the nature of the observed objects, as well as their conformation, thus ruling out chance or mere coincidence, as what was observed rather responds to premeditated planning and design in the laboratory.
Sommaire
Pictures Analysis
Zhao’s Research. DNA Tensegrity
DNA and graphene-based nanotechnologies
Boolean operations and logic gates with DNA
The CORDIS project of the DISCORDE program
Implications of the results
Evidence to comes
*************
Pictures Analysis
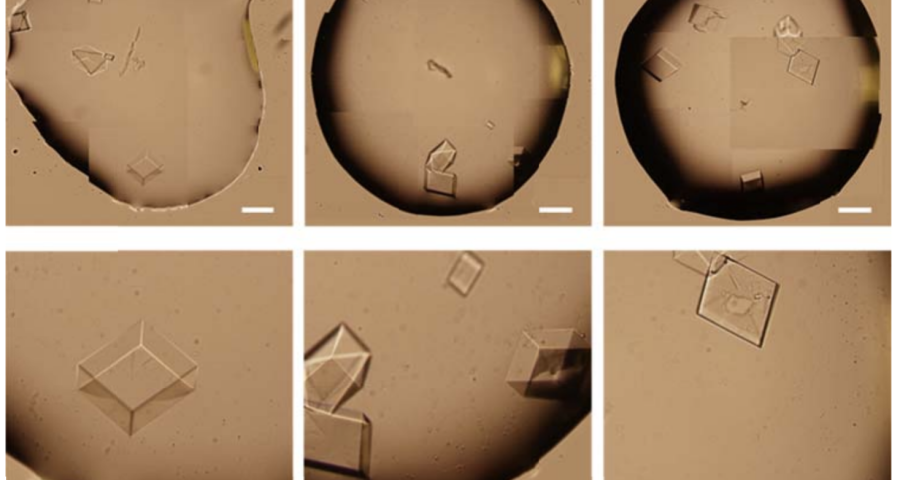
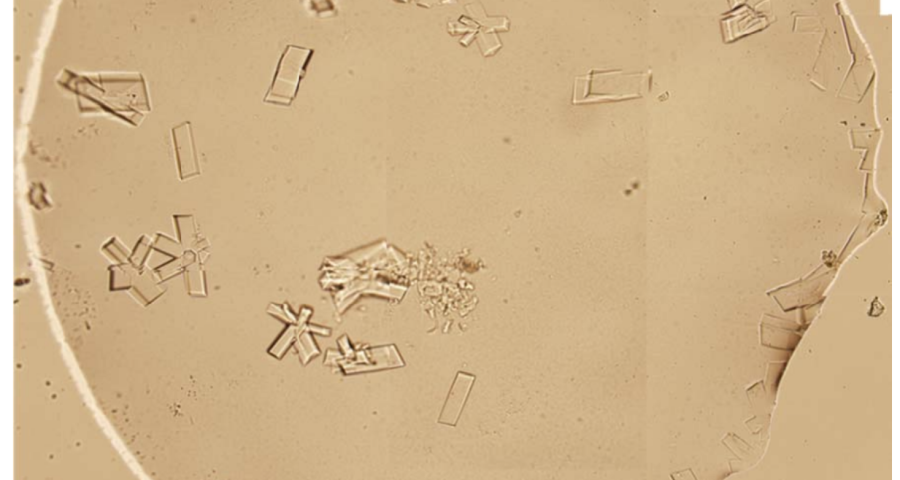
Figure 1 shows the sequence of images from the study by (Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018) in which the process of self-assembly of DNA crystals is observed at a scale of 200 μm – consistent with Dr. Campra’s observation scale. We can see how the “DNA molecules” form into two-dimensional, and three-dimensional crystals – with a quadrangular, rectangular, cubic structure, according to a predetermined order of construction. When the morphology of these DNA crystals is compared to the objects observed in Pfizer’s vaccine samples, the conclusion is that the match is exact – both in terms of formats, perspectives, dimensions and geometries.
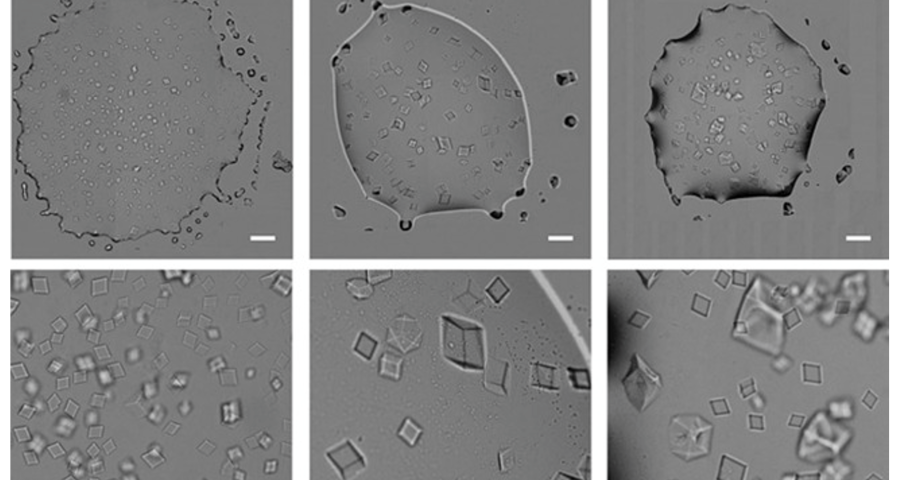
Figure 2 compares the images from Zhao’s study and those obtained by Dr. Campra, in his observation of the Pfizer vaccine, where it is clear that the objects observed are DNA crystals. Other references in the scientific literature also support this identification (Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018 | Chandrasekaran, A.R. 2019 | Hernandez, C.; Birktoft, J.J.; Ohayon, Y.P.; Chandrasekaran, A.R.; Abdallah, H.; Sha, R.; Seeman, N.C. 2017).
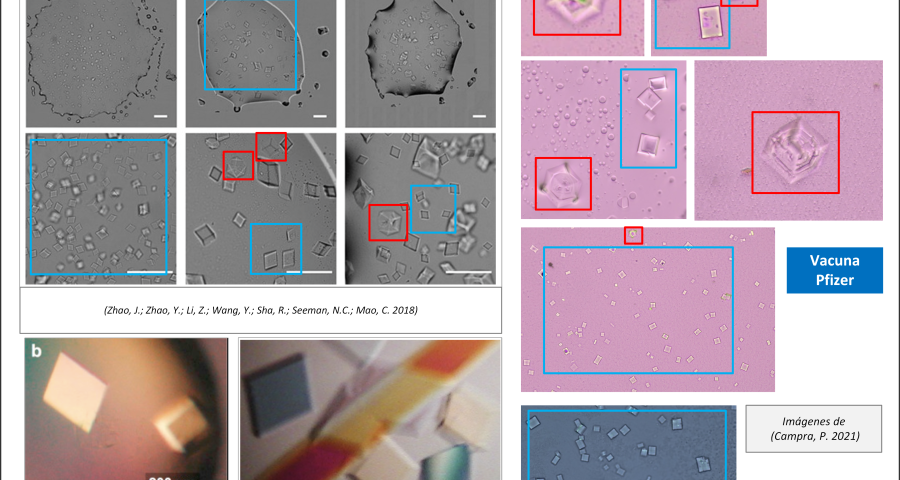
In addition, the recording of microscopic observations of the vaccine samples, by Dr. Campra and Ricardo Delgado, corroborated the presence of the DNA crystals and their assembly process, which corresponds to that described by Zhao.
In addition to these tests, Dr. Campra has prepared a document providing additional images consistent with the presence of DNA crystals in COVID-19 vaccines. This document can be viewed on his ResearchGate page. – or with the following permanent link.
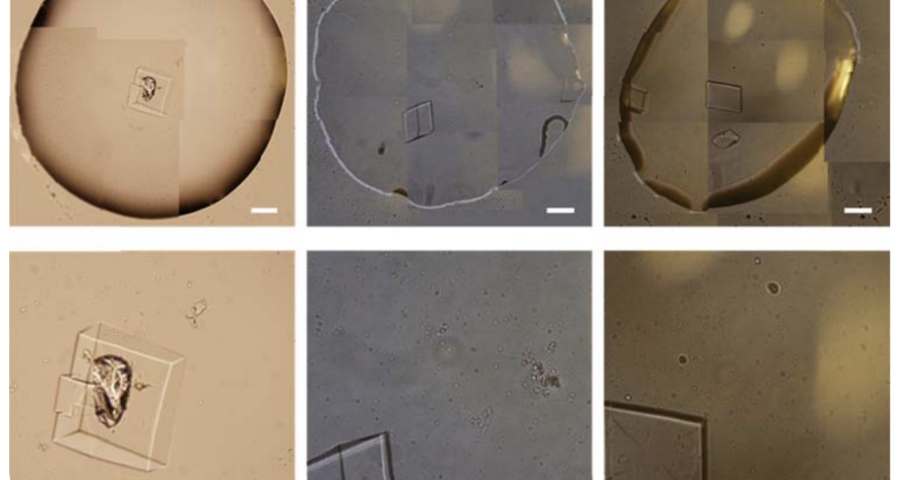
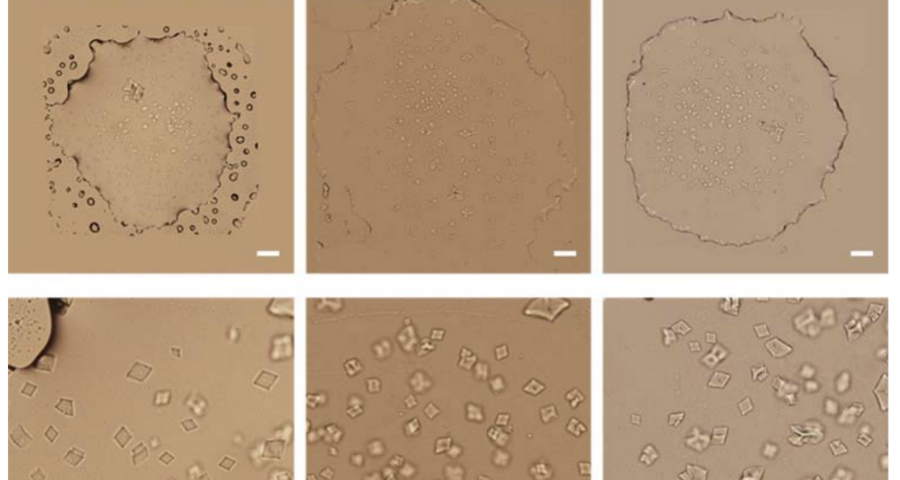
Additional evidence is presented in Figures 3, 4, and 5 below. For example, Figure 3 shows the presence of DNA crystals and what appears to be a carbon fiber or nanotube – which in reality could correspond to the structure of a DNA micro/nanotube. The significance of this discovery is enormous, as it involves reconsidering the composition of the objects identified so far, particularly the rectangular, rhomboidal and quadrangular structures and what appeared to be carbon nanotubes. Because of the complexity and the considerations, which arise for the research, this question will be the subject of a specific section in order to update the knowledge on these objects – explaining the current state of knowledge, at the time of writing this article.
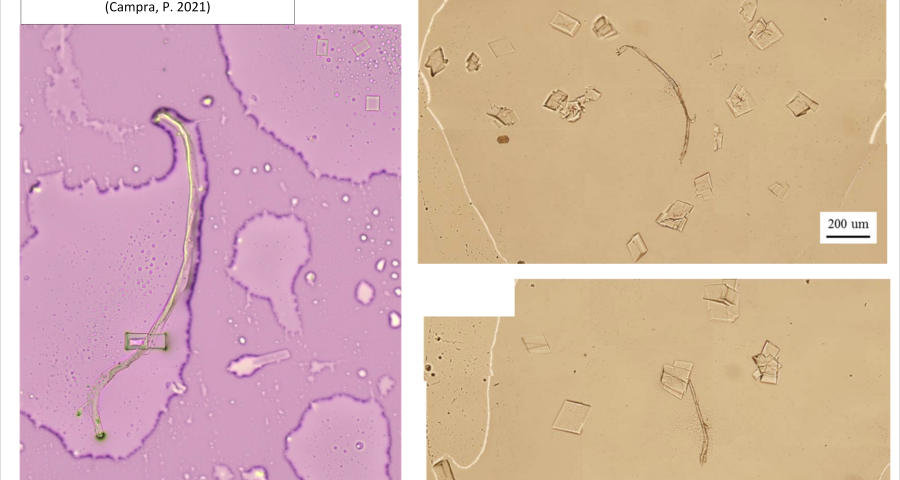
Figure 4 shows cubic structures reminiscent of clustered cubic pyrites – which would actually be the structures of the 3D DNA crystals being formed. It is interesting to note that the “cracks”, or “marks”, on the surface of these crystals are also very similar to those observed on DNA crystals in the scientific literature. Work is still in progress to find out what these cracks correspond to: whether they are construction defects or the result of the design of predetermined channels in the DNA molecules, or whether they follow a circuit pattern – as explained in the article on nanorods. The hypotheses are open and further research and studies are still needed to be able to make a more precise statement.
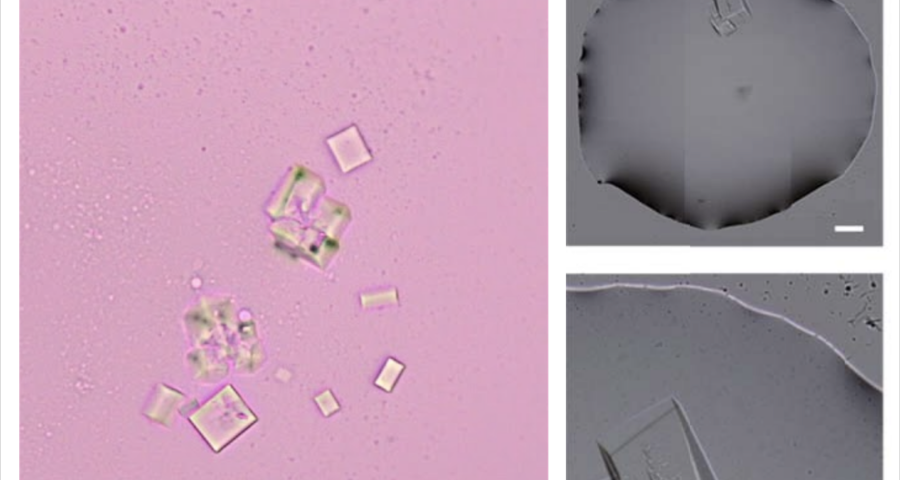
Figure 5 shows a fiber or micro/nanotube very similar, almost identical, to those observed in the vaccine samples, and in this particular case, from Pfizer. It is fair and honest to acknowledge that it is possible that many of the fibers observed in the vaccines do not correspond to single or compound wall carbon nanotubes, and are in fact single or compound wall DNA nanotubes, but most likely functionalized or hybridized with graphene, as will be explained, later, in the section on implications for the research conducted by Corona2Inspect.
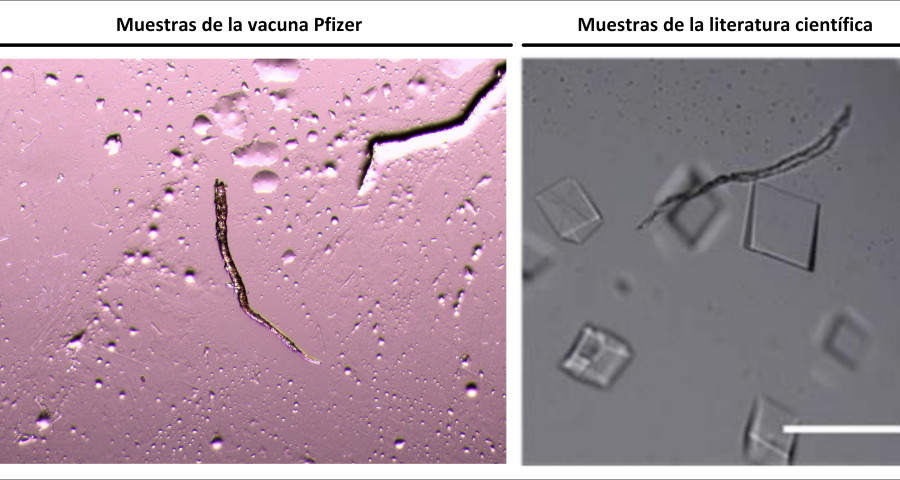
The importance of this discovery is paramount, as it changes many aspects of the paradigm and ecosystem of objects discovered so far in the vaccine images at a conceptual level. In addition to graphene and hydrogel, there would also be DNA used to build the observed structures, according to predetermined parameters, designs, patterns. This, therefore, leaves no doubt about the existence of nanotechnology and bioengineering, as explained in the work of (Zhao, J. Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018) and in the rest of the references of the scientific literature – which is very vast, in this field, and whose analysis and revision will take time. Other notable images of Zhao’s work are shown in Figures 6, and following.
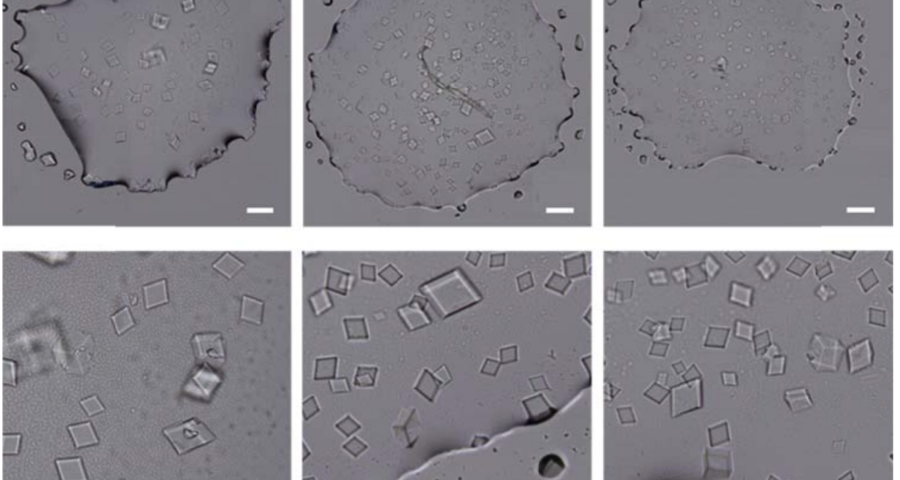
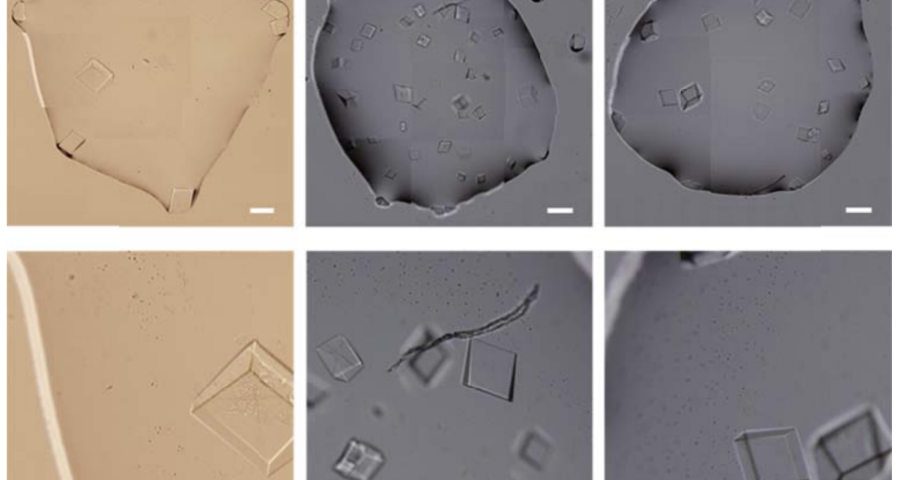
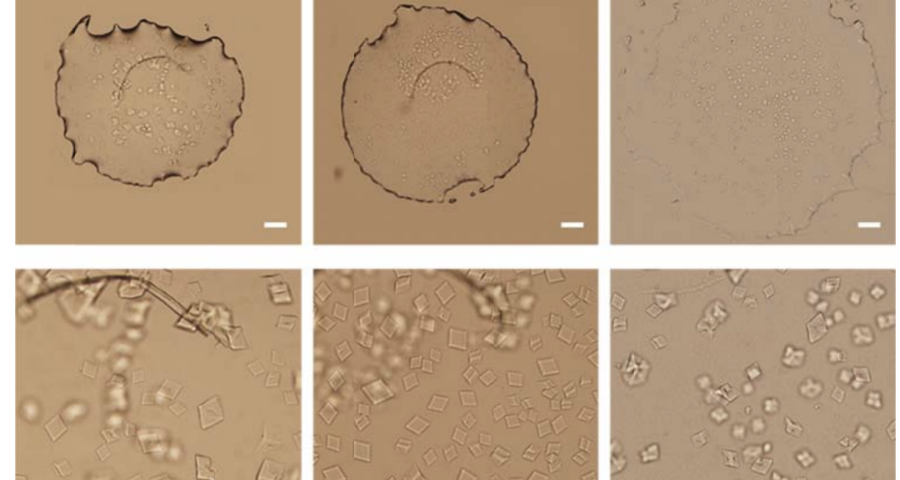
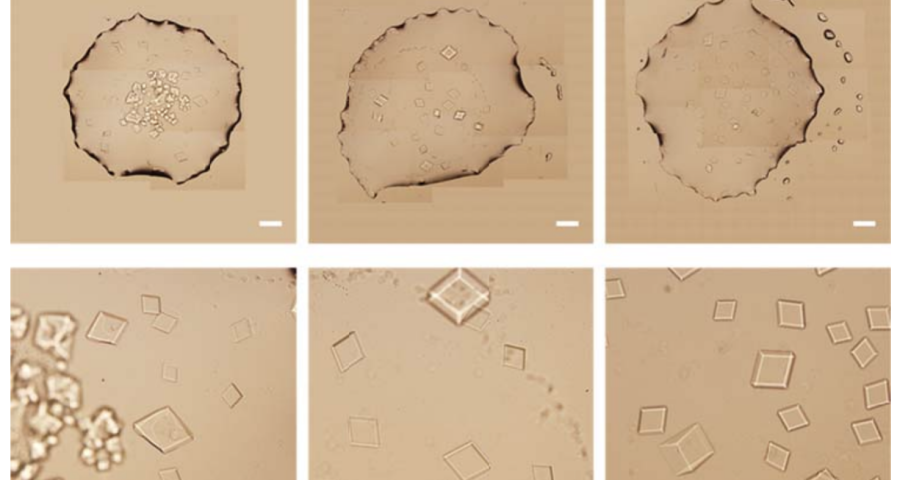

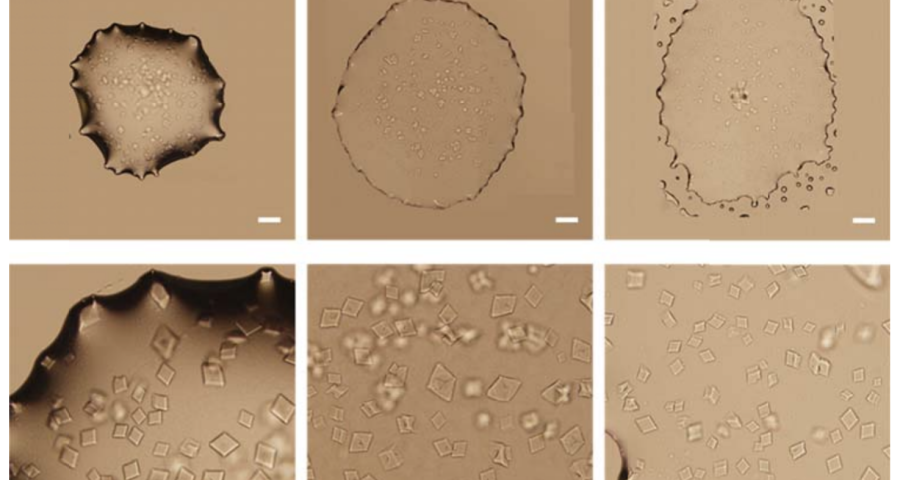

The rest of the images from Zhao’s research, and his team’s, can be found in the backgrounder to his scientific paper “Modulating Self-Assembly of DNA Crystals with Rationally Designed Agents” or at the following permanent link: [579]
*************
Zhao’s Research. DNA Tensegrity
The formation of DNA crystals is important for the development of nanotechnologies compatible with the human body but, also, to enable precise self-assembly processes to configure more complex devices. Indeed, «DNA is a powerful molecule for programmed self-assembly… Over the past decade, rationally designed 3D DNA crystals have been developed that can serve as scaffolds for enzyme loading and structure determination, filtration devices, and controllable device systems (Hao, Y.; Kristiansen, M.; Sha, R.; Birktoft, J.J.; Hernandez, C.; Mao, C.; Seeman, N.C. 2017)… In these crystals, the interactions between motifs (sticky-ended hybridizations) are rationally designed and can be easily programmed/modified based on Watson-Crick base pairing… Because of the programmability of DNA, these artificial DNA crystals are an excellent model system to study fundamental issues of bio-macromolecular crystallization». (Zhao, J. Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018). This makes DNA the ideal material to be administered in an aqueous solution, such as that of the vaccine.
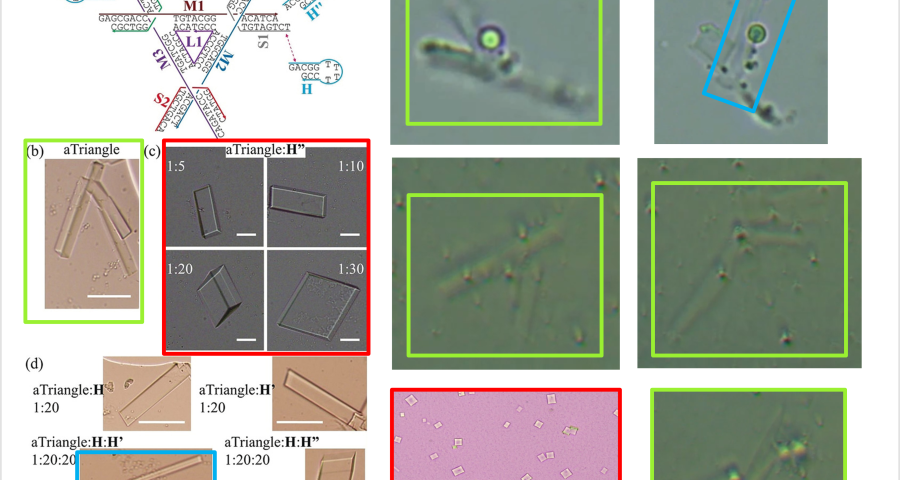
According to Zhao and his team, the creation of DNA crystals depends on the amount of crystallization nuclei – which are DNA molecules with a “DNA tensegrity triangle” structure – as well as the time factor. According to their study, «crystal growth depends critically on crystallization kinetics. In general, few nuclei lead to few (and large) crystals; a slow growth rate leads to high crystal quality. With the triangular DNA tensegrity crystal designed as a model system, we hypothesized that we could grow large crystals by adding an agent that interferes with crystal nucleation and growth. » The DNA tensegrity triangle appears to be an important element in the conformation of crystal structures, configuring a complex DNA lattice into predetermined geometric patterns – see Figure 19. The concept of “Tensegrity” refers to the structural principle that allows objects to be bound together in a taut network – which is applicable to the DNA strands that give rise to these crystals. In other words, the balance of the crystal depends on the tension exerted between the parts of the DNA chain that are in contact – this is especially true at the ends of the genome strands – at their junction with the rest of the DNA molecules.

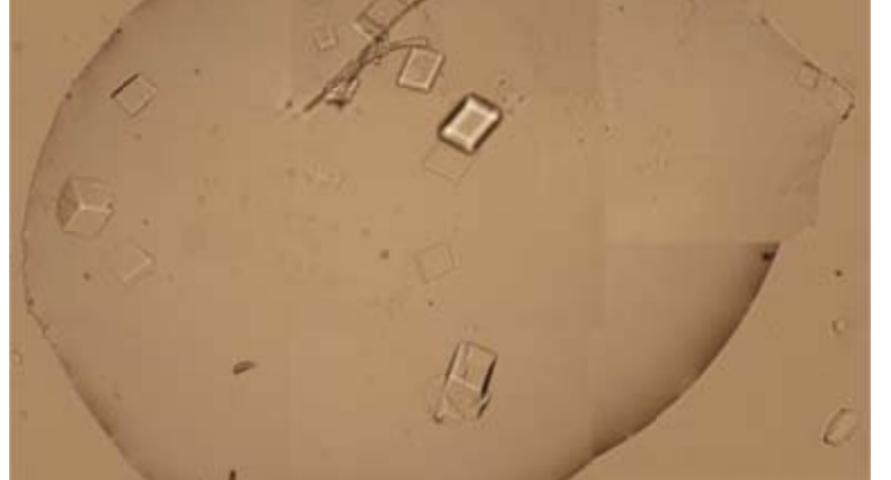
Furthermore, it is explained that the binding mechanism between DNA tensegrity triangles, or other DNA structures, is constituted by the ends of the genome strands and their nucleotides, which allows for the growth, orientation, dimensionality, and assembly constraint of crystals. The explanation is: «The basic pattern of DNA is a tensegrity triangle with triple rotational symmetry. There is a pair of complementary sticky ends, along each edge of the triangle, so that the triangular patterns can be associated with each other in three independent directions by an identical association of sticky ends to form 3D crystals. For this system, we designed a hairpin strand (H) of 12 nucleotides (nt) in length. It contains a 2 nt long sticky end, which is identical to the three sticky ends and complementary to the other three in the DNA triangle. The H-chain can transiently bind to the surface of the growing triangular crystals and the triangular motifs, and prevent the crystals from continuing to grow and prevent the triangular motifs from associating with each other.» This would demonstrate the control of crystal production, as well as the use of H-strands (hairpin) in the Pfizer vaccine, achieving identical results, as shown in the images in Figure 2, 20 and Video 1.
Other forms of DNA crystals are visible in Figure 20, depending on the H-strands, which act as modulating agents. Comparison with light microscopy images, of Pfizer vaccine samples, again shows great similarity. See also Figure 18.
*************
DNA and graphene-based nanotechnologies
Considering that DNA-based self-assembly technology, which forms crystals identical to those observed in Pfizer’s vaccine samples, could be considered to exist as well. This is revealed in the review of work by (Scalise, D.; Schulman, R. 2019). This is made possible by synthetic DNA strands, which use very specific sequences, that «can sense information in their environment and control the assembly, disassembly and reconfiguration of the material. These sequences could serve as inputs and outputs for DNA computer circuits, allowing them to act as chemical information processors to program complex behaviors in chemical and material systems.» In fact, they could not only process chemical information, as it is also added that «Specifically, there are interfaces that can release DNA strands in response to chemical, light wavelength, pH, or electrical signals, as well as DNA strands that can direct the self-assembly and dynamic reconfiguration of DNA nanostructures, regulate particle assemblies, control encapsulation, and manipulate materials including DNA crystals, hydrogels and vesicles. These interfaces could enable chemical circuits to exert algorithmic control over reactive materials, which could lead to the development of materials that grow, heal and interact dynamically with their environment.»
These statements by Scalise and Schulman are very illuminating, as they confirm that synthetic DNA structures can be used to create circuits, signal the wavelength of light, develop optoelectronic devices, and operate with electrical signals. In addition, it clearly refers to DNA crystals, hydrogels and vesicles that can be manipulated to develop controlled self-assembly and even self-healing. However, DNA circuits would not only be capable of operating with electrical signals. This is where graphene comes in, as it has been shown that it can be synthesized into the aptamers of DNA strands, giving them superconducting and electromagnetic signal-receiving capabilities, given their ability to absorb radiation and thus microwaves. This is stated by (Wang, L.; Zhu, J.; Han, L.; Jin, L.; Zhu, C.; Wang, E.; Dong, S. 2012) who developed a method to synthesize GO graphene oxide on DNA aptamers to create logic gates and multiplexing in the context of DNA circuit development. This is reflected in the following textual paragraph «a GO/aptamer system was constructed to create multiplexed logic operations and enable multiplexed target detection. The 6-carboxyfluorescein (FAM)-labeled adenosine triphosphate (ABA)-binding aptamer and the FAM-labeled thrombin-binding aptamer (TBA) were first adsorbed onto graphene oxide (GO) to form a GO/aptamer complex, resulting in FAM fluorescence quenching. We showed that the unique GO/aptamer interaction and target-specific recognition of the aptamer in the GO/aptamer/target system were programmable and could be used to regulate FAM fluorescence through OR and INHIBIT logic gates.» Configuring the aptamers with graphene oxide increases the electrical conductivity of the target DNA frame and allows them to act as field transistors, reducing the power consumption required to operate the intracorporeal nanocommunications network.
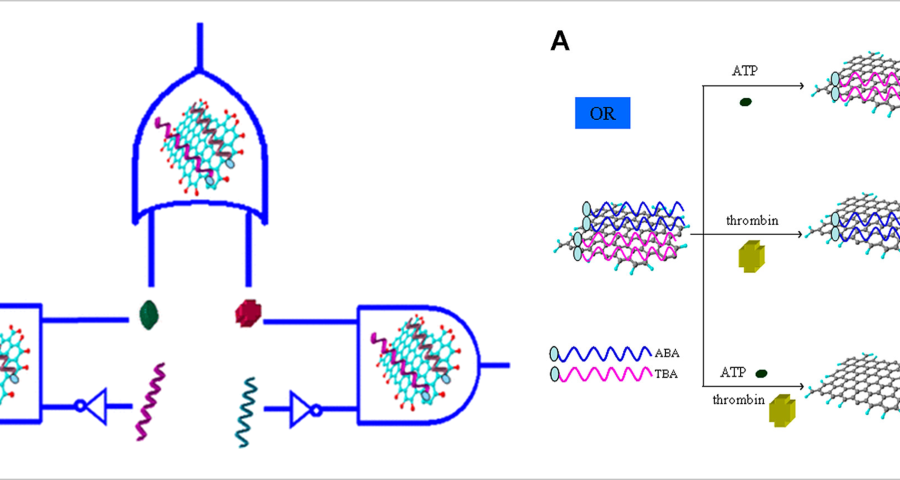
Going back to the work of (Scalise, D.; Schulman, R. 2019), it is repeatedly confirmed that the development of DNA circuits allows for the same operations as electronic transistor circuits, explicitly referring to this issue as follows «DNA circuits are a particularly promising medium for computation within chemical systems. They are primarily composed of DNA oligonucleotides (i.e., short strands of DNA), but may also contain enzymes such as DNA polymerase or exonucleases (Zhang, D.; Seelig, D. 2011 | Baccouche, A.; Montagne, K.; Padirac, A.; Fujii, T.; Rondelez, Y. 2014 | Willner, I.; Shlyahovsky, B.; Zayats, M.; Willner, B. 2008). DNA circuits can perform the same fundamental operations as transistor-based electronic circuits, including Boolean logic and arithmetic (6-9), oscillation generation and synchronization (10-13), and execution of interactive algorithms.» Based on these statements, which are widely documented in the scientific literature, there seems to be no doubt that traditional electronics, based on CMOS and silicon technology, is perfectly reproducible on a genetic scale. Therefore, there is no reason to doubt the presence of micro/nano electronic devices based on synthetic DNA. On the contrary, all the clues that have been explained in the course of this blog’s research point to the fact that vaccines, and Pfizer in particular, possess undeclared nanotechnology.
Scalise and Schulman explain very clearly how the input and output of this type of circuit works: «The inputs of DNA circuits are strands of DNA with specific sequences that can pass information about a material or environment to the circuit. Similarly, the outputs of a DNA circuit are strands that can control the states of materials or molecules. The input/output strands of DNA circuits are thus analogous to USB ports on electronic computers, i.e., a standard interface that allows the circuit to communicate with peripherals, in this case molecules or materials. The use of modular input/output interfaces is a key design principle that allows the same types of circuits to interact with a diverse set of materials. In principle, an input sensor can be replaced by another type of sensor to allow the same circuit to receive and process information about a different type of environmental stimulus. Similarly, output actuators can be swapped to allow the same circuit to drive different responses.» This provides insight into the interoperability of DNA circuits, with well-defined data ports.
The development of self-assembled DNA molecular circuits would be done in an appropriate medium, namely by the use of hydrogels. This is important because the presence of hydrogels in vaccines had been discussed, as explained by the entry on the 1450 Raman spectrum detected in vaccine vials, according to Dr. Campra’s result. In fact, Scalise and Schulman state that «hydrogels are materials composed of polymer chains cross-linked in water. By incorporating DNA into the hydrogel in the form of cross-links (Nagahara, S.; Matsuda, T. 1996), the material properties of the hydrogel can be dynamically manipulated by adding DNA strands to modify, break, or create cross-links. According to (Lin, D.C.; Yurke, B.; Langrana, N.A. 2004), they developed DNA crosslinks that could be reversibly dissociated by adding a complementary strand to one of the strands in the crosslink. DNA-crosslinked hydrogels can also be stiffened or softened by the addition of DNA that changes the conformation of the crosslinks between a double-stranded state or a partially single-stranded conformation (Lin, D.C.; Yurke, B.; Langrana, N.A. 2005) by DNA strand displacement reactions.» This description is consistent with DNA crystals, which initially appear flexible during the self-assembly process (see Video 1), but stiffen at the end of their construction, exhibiting a coherent or rigid morphology.
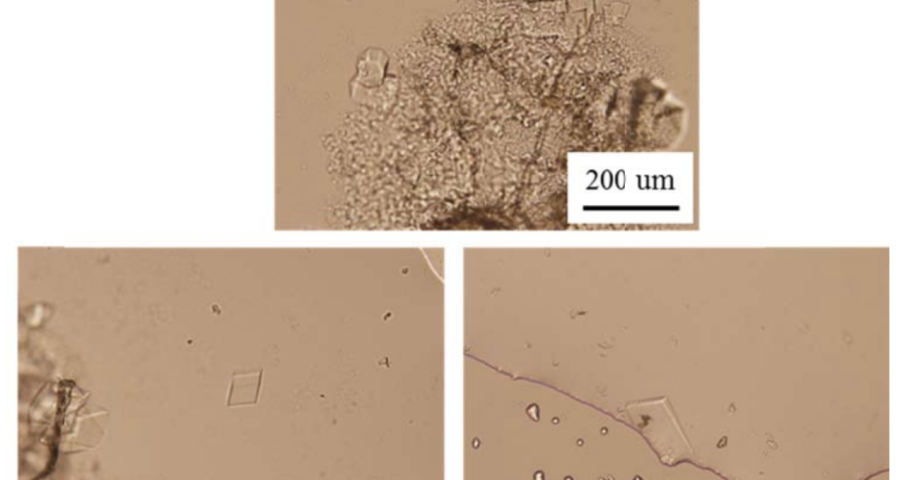
Another detail that would confirm that what is observed in the microscopy images are traces of fluorescent staining, which would develop in three-dimensional structures, following the modification or alteration of their DNA strands or the introduction of colored oligonucleotides. In the words of (Scalise, D.; Schulman, R. 2019 | Hao, Y.; Kristiansen, M.; Sha, R.; Birktoft, J.J.; Hernandez, C.; Mao, C.; Seeman, N.C. 2017), it is stated that «DNA strand displacement reactions operate within three-dimensional DNA crystals, for example, using DNA strands with different fluorescent modifications to change the color of a crystal.» This staining practice is reported in several papers, among which it is worth highlighting that of (Rusling, D.A.; Chandrasekaran, A.R.; Ohayon, Y.P.; Brown, T.; Fox, K.R.; Sha, R.; Seeman, N.C. 2014) where a fluorescent teal stain is obtained, similar to some of the traces observed in the crystals of the photographed samples of the Pfizer vaccine, see Figure 22.
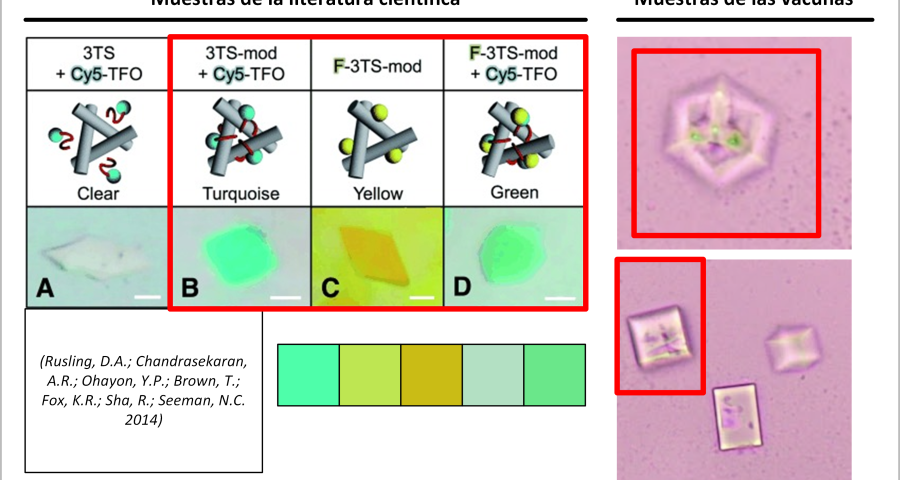
Furthermore, Rusling and his team’s research is very much in line with what has already been described in this post, stating that «DNA is a very useful molecule for programmed self-assembly of 2D and 3D nanoscale objects. The design of these structures takes advantage of Watson-Crick hybridization and strand exchange to join linear duplexes into finite assemblies. The dimensions of these complexes can be increased by more than five orders of magnitude by self-assembly of single-stranded cohesive segments (cohesive ends). Methods that exploit the sequence addressability of DNA nanostructures will enable programmable positioning of components in 2D and 3D space, offering applications such as organizing nanoelectronics, directing biological cascades, and determining the structure of periodically positioned molecules by X-ray diffraction. To this end, we present a macroscopic 3D crystal based on the rotationally symmetric triple tensegrity triangle, which can be functionalized with a triplex-forming oligonucleotide on each of its helical edges (TFO)… In the presence of TFO, the crystals were green, further demonstrating the successful incorporation of TFO into the crystal.»
Regarding the possibility of developing electronic circuits and devices, there is an abundant scientific literature demonstrating the direct implication of self-assembled DNA to create computer systems and nanotechnologies, also by means of DNA crystals. An example of this can be found in the article by (Jackson, T.; Fitzgerald, R.; Miller, D.K.; Khisamutdinov, E.F. 2021) which literally explains the following: «The integration of advances in nucleic acid nanotechnology and nucleic acid aptamer technologies enables the construction of novel nanoparticles that perform intermediate functions between electronic computers and biological systems. Programming with biological molecules, especially nucleic acids (NAs), is becoming very attractive because of their potential for functions ranging from simple fluorescence emission to sophisticated in vivo gene regulation. The structural behavior encompassed by their sequences can be predicted and manipulated by 2D folding algorithms. The resulting nucleic acid biopolymers can be used as logic-driven nanoagents for specific biomedical applications. Fluorogenic RNA aptamers can be designed to function as a single circuit within individual binary logic gates. This demonstrates the great potential of nucleic acid nanotechnology and promises to develop cutting-edge technologies, especially if combined synergistically with other computing and nanorobotic systems.»
More importantly, the scientific research confirms that the devices could be designed for “signal transduction and processing” which overlaps with the tasks performed by routers, transducers, transducers, controllers, explicitly mentioned in the context of the intracorporeal wireless nanocommunications network. Indeed, Jackson and his team state that «DNA nanotechnology could provide a simple approach to modularly design user-friendly interfaces for signal input/output, in which all transduction, information processing and signal generation tasks are performed by DNA/RNA circuit elements. First, because many DNA nanostructures are easily internalized by living cells and possess numerous binding sites, they can be used as multifunctional vectors to deliver regulatory proteins and DNA/RNA circuitry to living organisms. In addition, these complexes can be guided to specific organelles or organs using small ligands, aptamers, or peptide signals preloaded onto DNA nanostructures. These systems would enable the design of a universal system to deliver genetic instructions to the cell using modules based on DNA nanostructures. Second, dynamic DNA nanostructures incorporating FNAs (functional nucleic acids) offer great flexibility in the design of signal transducers due to the availability of a wide range of FNAs identified by SELEX (Systematic Ligand Evolution by Exponential Enrichment) or extracted from natural genomes. These transducers can specifically transform an input signal into digital binary states (On 1/Off 0) by structural switching. Regulatory biomolecules released during structural switching can be relayed and reconfigured to intracellular signaling pathways or circuits. Third, DNA/RNA circuits can act directly on nucleic acids produced during input signal transduction to process information and make logical decisions. Fourth, once signals have been processed, DNA/RNA can be used to generate output signals that are transmitted to an external observer in other cells.»
*************
Boolean operations and logic gates with DNA
The abundance of scientific literature on the design of Boolean arithmetic, computation, logic gates with DNA, among others, could be described as exceptional. It is very prolific, see some references (George, A.K.; Kunnummal, I.O.; Alazzawi, L.; Singh, H. 2020 | Fan, D.; Wang, J.; Wang, E.; Dong, S. 2020 | Xiong, X.; Xiao, M.; Lai, W.; Li, Li. Lai, W.; Li, L.; Fan, C.; Pei, H. 2021 | Zhao, S.; Yu, L.; Yang, S.; Tang, X.; Chang, K.; Chen, M. 2021 | Yao, C.Y.; Lin, H.Y.; Crory, H. S.; de-Silva, A.P. 2020 | Zhou, Z. Wang, J.; Levine, R.D.; Remacle, F.; Willner, I. 2021 | Katz, E. 2020 | Lv, H.; Li, Q.; Shi, J.; Fan, C. Wang, F. 2021 | Jiang, C.; Zhang, Y.; Wang, F.; Liu, H. 2021 | Liu, Q.; Zhang, Y.; Wang, F.; Liu, H. 2021 | Liu, Q. Yang, K.; Xie, J.; Sun, Y. 2021 | Chen, Z.; Yin, Z.; Cui, J.; Yang, J.; Tang, Z. 2021 | Zhang, J.; Liu, C. 2021 | Wang, Y.; Qian, M.; Hu, W.; Wang, L.; Dong, Y. 2020). Most research agrees that DNA strand displacement is the property that enables the development of molecular computing models. Thus, digital switching circuits, logic gates, flow switches, allow conventional electronics to be adapted to the digital molecular computing model. This is confirmed, for example, by (Chatterjee, G.; Dalchau, N.; Muscat, R.A.; Phillips, A.; Seelig, G. 2017) in their paper “A spatially localized architecture for fast modular DNA computing”, which was the cover story of the Microsoft research blog announcing the design and production of nanoscale computing circuits with DNA. This is shown in the work of (Wang, F.; Lv, H.; Li, Q.; Li, J.; Zhang, X.; Shi, J.; Fan, C. 2020) where we can see how the activation and deactivation of genetic sequences are used to perform arithmetic computations, see figures 23 and 24. This is literally extrapolable to operation with binary code and thus to the operation of transistors and integrated circuits, see some examples (Polonsky, S.; Stolovitzky, G.; Rossnagel, S. 2007; Ogata, N. 2012; Gupta, R.K.; Saraf, V. 2009; Sawlekar, R.; Nikolakopoulos, R.; Nikolakopoulos, C. 2020). Yan, S.; Wong, K.C. 2021 | Bhalla, V.; Bajpai, R.P.; Bharadwaj, L. M. 2003 | Zahid, M.; Kim, B.; Hussain, R. Amin, R.; Park, S. H. 2013 | Matsuo, N.; Takagi, S.; Yamana, K.; Heya, A.; Takada, T.; Yokoyama, S. 2012 | Lyshevski, MA 2005 | Wang, K. 2018).


*************
The CORDIS project of the DISCORDE program
In spite of the irony and the license I decided to take in the title of this section, I consider it fair to indicate “DISCORDE” in a pun, which allows us to attend the following CORDIS project with the identifier 321772, entitled “Logical re-routing of cellular communication networks by DNA origami nanorobot – Final report summary – DNA NANO-ROUTERS”, see (CORDIS. European Commission. 2019). The title is explicit enough to confirm that it is possible to develop self-assembled DNA origami nanorouters that develop logical computing. This confirms the indications of nanorouters in an intracorporeal network of nanocommunications and nanodevices in the human body, as explained in Corona2Inspect. In fact, the CORDIS project abstract literally states the following:
«The goal of the nanorouters project was to design nanoscale robots made from synthetic DNA molecules capable of redirecting cell-to-cell communication, and demonstrate this in a clinically relevant model….. The work carried out in the project focused on designing large-scale synthesizable DNA robots made entirely of DNA (and without including chemical drugs or proteins, in order to maintain material homogeneity and manufacturing simplicity); demonstrating them in multiple cell-like systems in vitro and, finally, demonstrating them in-vivo, was completed in late 2017.».
From this statement in the final report of the project, it is deduced that nanowinders exist to enable signal and data routing in the context of cellular, molecular and thus intracorporeal nanocommunication networks, thus addressing the need to develop and test nanotechnologies that would then be introduced into vaccines.
*************
Implication of the results
The presence of self-assembled DNA crystals in vaccine samples has important implications for ongoing research. First, their mere presence in the vaccines implies the use of highly advanced nanotechnologies, the self-assembly of objects according to predetermined patterns and templates in the laboratory, and thus confirms premeditated intentionality and purpose, with the clear and obvious fact that there is synthetic artificial DNA, which together with graphene constitutes a new and undeclared element in the vaccine. Secondly, this discovery has important implications for the lines of research that deal with the study and analysis of vaccines, since there is no longer just graphene or a hydrogel available. We now have to consider the presence of all kinds of self-assembled objects with DNA, which expands the possibilities of bioengineering in the human body, the development of biosensors and devices in the context of the intracorporeal wireless nanocommunications network. Third, the presence of DNA crystals has been demonstrated and therefore implies, with high probability, the presence of DNA-based circuits and computer systems. As has been demonstrated, there is ample scientific evidence that it is possible to develop circuits, transistors and electronic devices with DNA strands, allowing the operation of all kinds of signals and data, including binary type.
Carbon nano-tubes and DNA nano-tubes
From Figures 3, 5, 7, 10, among others observed in the work of (Zhao, J.; Zhao, Y.; Li, Z.; Wang, Y.; Sha, R.; Seeman, N.C.; Mao, C. 2018), it is concluded that some of the fiber-like objects may not actually correspond to CNT micro/carbon nanotubes, being in fact DNA micro/nanotubes. However, it still cannot be ruled out that carbon nanotubes do not exist in vaccines as a result of folding graphene sheets or nanofiches. Indeed, the presence of graphene oxide in vaccines implies, with high probability, the presence of carbon nanotubes at one or more walls. It is therefore plausible that there are both carbon and DNA micro/nanotubular structures intermingled in the vaccine solution. It is also important to note that the scientific literature clearly and convincingly indicates that DNA can hybridize, functionalize, or decorate with graphene in aqueous solutions (Sun, J.; Li, Y.; Lin, J. 2017 | Premkumar, T.; Geckeler, K.E. 2012 | Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. 2018 | Zhang, H.; Gruener, G.; Zhao, Y. 2013), being most likely the presence of DNA micro/nanotubes and graphene, a hybrid material that would obtain the superconducting, electromagnetic, piezoelectric properties of CNT carbon nanotubes, which would be easy to assimilate into the target organs of the human body since it is made of DNA. In other words, the release of micro/nanotubes would be minimized (Gangrade, A.; Stephanopoulos, N.; Bhatia, D. 2021 | Fu, X.; Peng, F.; Lee, J.; Yang, Q.; Zhang, F.; Xiong, M.; Zhang, X.B. 2020 ) to enter the central nervous system and cardiac tissues, preferably (Sidharthan, D.S.; Abhinandan, R.; Balagangadharan, K.; Selvamurugan, N. 2021 | Bhattacharya, S. 2021). Indeed, there are clear references around the development of DNA nano-biosensors, carbon nanotubes and graphene quantum dots (Qian, Z.S.; Shan, X.Y. Chai, L.J.; Ma, J.J.; Chen, J.R.; Feng, H. 2014 | Chi, Q.; Yang, Z.; Xu, K.; Wang, C.; Liang, H. 2020 | Tam, D.Y.; Ho, J. W.T.; Chan, M.S.; Lau, C.H.; Chang, T.J.H. Leung, H.M.; Lo, P.K. 2020 | Stephanopoulos, N.; Freeman, R.; North, H.A. ; Sur, S.; Jeong, S.J.; Tantakitti, F.; Stupp, S.I. 2015 | Silva, G.A.; Khraiche, M.L. 2013), but also DNA aptamer hybridization (Wang, L.; Wu, A.; Wei, G. 2018 | Wang, L.; Zhu, J.; Han, L.; Jin, L.; Zhu, C.; Wang, E.; Dong, S. 2012).
Transistors and DNA circuits, microfluidic pipelines, QCA circuits
According to all the elements already observed in the microscopic images of the vaccines, according to the scientific literature and assuming the presence of synthetic DNA structures, we can deduce that the rectangular, quadrangular structures, more or less complex in their design, which look like circuits, are in reality, what we observe in appearance, integrated circuits. Indeed, according to (Mohanty, N.; Berry, V. 2008 | Green, N.S.; Norton, M.L. 2015 | Premkumar, T.; Geckeler, K.E. 2012), among others, the development of transistors, bio-devices, biosensors and integrated circuits based on DNA and graphene is feasible. Regarding the detection or presence of microfluidic channels, referred to as microfluidic chips (Materón, E.M.; Lima, R.S.; Joshi, N.; Shimizu, F.M.; Oliveira, O.N. 2019 | Ang, P.K.; Li, A.; Jaiswal, M.; Wang, Y.; Hou, H.W.; Thong, J.T.; Loh, K.P. 2011), in some of the objects observed in vaccines, is a possibility that cannot be ruled out yet. Indeed, crystals and DNA structures would not be incompatible with the design of such pipelined chips and circuits. However, there are some problems with this approach. In most cases, microfluidic circuits appear in the scientific literature at a much larger scale than that observed in vaccine samples. While it is not impossible to find papers reporting microfluidic designs at the micrometer scale, it is true that they generally require relatively conventional processing and analysis techniques using microscopy, camera, and computer equipment for direct analysis. On the other hand, logically, if it is possible to create circuits and transistors with DNA and graphene, it would not make much sense to build microfluidic chips much larger than the molecular scale. It is the principle of economy of means and limited resources that must be considered in any inoculation. Therefore, it does not seem obvious that what we are observing are microfluidic circuits, although they may appear to be. On the other hand, regarding the QCA (Quantum Cellular Automata) circuits described in the article on nanorods, although what has been observed suggests a quantum dot circuit composed of wires and logic gates, among other components, it is still a real and feasible possibility. In fact, there are scientific papers that demonstrate the real possibility of creating QCA molecular-scale circuits from DNA nanopatterns (Hu, W.; Sarveswaran, K.; Lieberman, M.; Bernstein, G.H. 2005 | Walus, K.; Karim, F. ; Ivanov, A. 2009 | Dysart, T.J. 2009 | Ma, X.; Lombardi, F. 2008 | Ma, X.; Lombardi, F. 2009 | Hänninen, I.; Takala, J. 2010 | Jang, B.; Kim, Y.B.; Lombardi, F. 2008), see also the work of (Chaudhary, A.; Chen, D.Z.; Hu, X.S.; Niemier, M.T.;
«The requirements for positioning QCA molecular cells are stringent. More than in any other implementation, individual molecules must be placed in specific locations with nanometer precision. Here, we will begin to explain exactly how precise it is necessary to make QCA circuits and why building the crossover will be physically challenging. Recent results on DNA mosaics suggest that DNA could be used as a circuit board to place QCA cells. A viable target involves DNA mosaics as a scaffold. The QCA molecules would covalently bind to the modified nucleotides in the DNA. Assuming that the mosaics retain the B-DNA duplex, sites in the main groove could serve as binding sites with a periodicity of 3.6 Å (angstrom unit of measurement) along the helix axis and 2 or 4 Å perpendicular to the helix axis (creating the potential for densities of 1013/cm2). This paper has significant critical mass, as many research groups are currently seeking to create and attach nanoparticles to DNA-based scaffolds.»
This explanation is highly indicative of the type of circuitry that could be found in vaccine samples. The authors make explicit the use of DNA that would act as a plate or scaffold for the deposition of QCAs to form a circuit. These QCAs could be made of different materials, including graphene, using graphene quantum dots (GQDs) or graphene quantum dots. Chaudhary also makes a direct connection to self-assembly, as shown in the following paragraph:
«DNA rafts containing molecules can be guided to certain locations to form more complex systems. Previously, we discussed the use of chemical synthesis and bottom-up self-assembly to form molecular circuit boards. This section discusses how these circuit boards will be used to form systems. Directed assembly envisions the use of top-down lithography from 10 to 100 nm to guide the bonding of printed circuits to CMOS substrates. Interactions between the printed circuits will allow them to couple with specific partners, while maintaining sub-nanometer registration from one printed circuit to the next».
For all these reasons, the hypothesis of QCA circuits such as the nano-turner cannot yet be ruled out, on the contrary, it is more likely to be found in the contents of the vaccines. However, the search is still open and new clues, evidence and testimonies can be found to reinforce the identifications that have been made. Readers should be reminded that the nature of the research being conducted is complex and follows the methodological model of reverse engineering. These are difficult investigations in which knowledge is established little by little, as new indications and evidence are integrated that allow rethinking, in some cases, aspects that could have been taken for granted. By virtue of what has been said, it cannot be excluded that the type of circuit used is simply synthetic DNA strands encapsulated in DNA crystals, to cite another case that has a high probability of being found.
DNA circuits in the intracorporeal wireless nanocommunications network
According to the work of (Zhang, Y.; Feng, Y.; Liang, Y.; Yang, J.; Zhang, C. 2021), it is possible to create “synthetic DNA circuits and networks for molecular information processing”, which corresponds to the type of communication, “molecular”, resulting from the functioning of neurons in the central nervous system, according to the scientific literature consulted (Balasubramaniam, S. ; Boyle, N.T. ; Della-Chiesa, A. ; Wals, F. ; Mardvoglu, A. ; Botvich, D.). Della-Chiesa, A.; Walsh, F.; Mardinoglu, A.; Botvich, D.; Prina-Mello, A. 2011 | Suzuki, J.; Budiman, H.; Carr, T.A.; DeBlois, J.H. 2013 | Abd-El-atty, S.M.; Lizos, K.A.; Gharsseldien, Z.M.; Tolba, A.; Makhadmeh, Z. A. 2018 | Yang, K.; Bi, D.; Deng, Y.; Zhang, R.; Rahman, M.M.U.; Ali, N.A.; Alomainy, A. 2020) and in line with the entry on intracorporeal nanocommunication networks and carbon nanotubes. In fact, Zhang and his team state that «through a DNA strand displacement reaction, a synthetic DNA circuit can be used to precisely regulate complex genetic networks and molecular biosystems, e.g. DNA neural network systems….. Synthetic DNA circuits have proven to be superior in simulating and regulating DNA signaling, due to their programmability and ease-of-use properties… A variety of bioengineering and bioinformatics functions have been regulated by varying the architectures and integrations of DNA circuits, such as simulation of their signals, molecular switches, catalytic cycles, cascade amplification and logic gates.» With this explanation, the relationship between the development of DNA circuits, DNA nanotubes and carbon (graphene) and the objective of serving the calculation and reception of signals and data resulting from molecular communication, which occurs mainly in the central nervous system and specifically in the neuronal tissue of the brain, becomes clear. We can therefore conclude that DNA provides the computational capacity of the artificial neural network that can be configured in the central nervous system, processing the signals generated at the molecular level by the neurons, transmitting and propagating the coded information to the rest of the network by means of micro/nano-antennas, micro/nano-rectennas with plasmonic properties.
Mass production of DNA structures
One of the questions that can be raised around the discovery of DNA crystals and artificial sequences to build structures and circuits is the consideration of mass and serial production of genetic sequences and their introduction into vaccines. This may seem like an impossible challenge, but science has managed to solve this problem, see the work by (Praetorius, F.; Kick, B.; Behler, K.L.; Honemann, M.N.; Weuster-Botz, D.; Dietz, H. 2017) titled “Biotechnological mass production of DNA origami”. In fact, he explains that the synthetic DNA origami used in self-assembly is designed from a «nanostructure from a very long single-stranded scaffold molecule held in place by numerous single-stranded base oligonucleotides» and goes on to state that «only bacteriophage-derived scaffold molecules are amenable to evolutionary and efficient mass production ; shorter short strands are obtained by solid-phase synthesis or by expensive enzymatic processes. …. we show that single strands of DNA of virtually arbitrary length and with virtually arbitrary sequences can be produced in an evolutionary and cost-effective manner using bacteriophages to generate single-stranded precursor DNA.» There is no doubt that mass production of synthetic DNA is feasible, both in quantity and efficiency, which is consistent with mass production of millions of vaccines.
Hybridization of synthetic DNA and graphene in the human body
Although research into the hybridization of synthetic DNA and graphene in the human body has not yet begun, clues that support this hypothesis are already beginning to emerge. The fact that graphene can combine and hybridize with synthetic DNA in vaccines, especially at the level of strand aptamers, allows one to infer, or even deduce, that it could also do so with the natural DNA of the human body. In the absence of relevant research and a review of the scientific literature, this possibility should be highlighted as being extremely concerning, given the consequences, purposes and sequelae it could have. Speculating on this approach, it is not inconceivable that DNA circuits could reproduce like the rest of the body’s DNA strings, which would require the CRISPR-Cas-9 or CRISPR-Cas-13 gene-editing technique, which would mean editing the human being’s genes, to transform it into an intelligent device, connected to the network, constituting a source of bio/neurological information and data, interactive and stimulated by any type of compatible signal received from the outside, if we take into account the principles of the intracorporeal network of nanocommunications. On the other hand, we must not forget the detail of the hybridization of graphene in the DNA aptamers that, by dividing and degrading into quantum dots, could contaminate the natural DNA and transform it into a superconductor, being particularly applicable in the nervous system of the human body, transforming it into a fiber optic highway through which signals and data are efficiently transmitted and propagated to the four corners of the body. Another very important consequence is the transmissibility of the genetic modifications that may have occurred, i.e. the “genetic contamination”, whose purification and reversibility are still unknown.
*************
Evidence to come
The importance of this discovery requires extraordinary tests that can be conclusively proven. To this end, two scientific tests are being developed to ensure the presence of DNA in the vaccine vials. On the other hand, a catalog of DNA sequences from the scientific articles consulted in relation to DNA crystals, DNA self-assembly, hybridization of aptamers and synthetic DNA, DNA loops, etc. has been elaborated. In this case, the objective will be to compare DNA strands collected from the specialized scientific literature and vaccines. Through these tests, we will try to find the definitive evidence that will allow us to state, without any margin of error, that the vaccines contain synthetic DNA, nanotechnology, self-assembly of nano-electronic components and devices, DNA circuits, etc.
*************